Table of contents
- Description of platforms.
- Sequencing services.
- Bioinformatics services.
- Access to services.
- High-throughput proteomics services.
- Terms and conditions of services.
- Cost of services.
- Institutional collaboration
1. Description of platforms
ITER’s Genomics Service has several second-generation (MiSeq, NextSeq 550 and NovaSeq 6000) and third-generation (MinION Mk1B/Mk1C and GridION x5) mass sequencing platforms that allow the sequencing of nucleic acids by short and long reads, respectively. In addition, during 2024, work is underway to acquire a high-throughput proteomics platform based on mass sequencing (Figure 1). All these services are provided for research (Research Use Only).
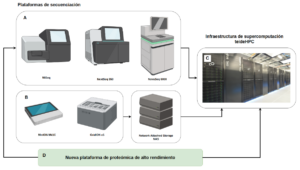
Figure 1. High-throughput genomics and proteomics platforms connected to the ITER supercomputing infrastructure.
These high-performance platforms, genomics and proteomics, are interconnected with ITER’s supercomputing infrastructure, teideHPC, which facilitates their time-bound storage and allows bioinformatics computational tasks to be carried out. To learn more about these infrastructures, click here (https://teidehpc.iter.es/). The HPC infrastructure services can also be accessed independently (https://teidehpc.iter.es/servicios/).
If you have already extracted nucleic acids (Figure 2), under the appropriate conditions, our team, through the so-called Genomics Service, has the technology for analysis from sample to interpreted result, offering accurate results at competitive prices adapted to the scale of your project. We also have the capacity to support large-scale projects.
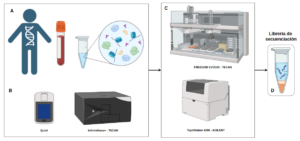
Figure 2. Schematic of the workflow from sample to preparation of nucleic acid libraries suitable for bulk sequencing on our platforms.
All these services are supervised in the Genomics Laboratory by a professional team with the appropriate scientific and technical qualifications. In addition, we are currently working on the implementation of the appropriate measures to apply for ENAC ISO 17025 accreditation for both whole genome and whole exome sequencing during the year 2025.

In our facilities we routinely offer the following services:
- Whole human genome sequencing (WGS).
- Whole human exome sequencing (WES).
- Other types of sequencing:
- RNA sequencing.
- Metagenomic or shotgun sequencing.
- ChIP-Seq.
- Methyl-Seq.
- Barcoding molecular (16S rRNA, COI)
- Sequencing of pathogens (viruses, bacteria).
- Bioinformatics services: primary, secondary and/or tertiary analysis.
If you require any other type of sequencing or would like to know if we can be of assistance in the analysis of the sequence data obtained, please contact our team.
2. Sequencing services.
2.1. Whole human genome sequencing (WGS)
- Coverage options: minimum 30x.
- Library preparation: Illumina DNA Prep or Illumina DNA PCR-Free Prep, among others.
- Human WGS quality controls.
- The sequenced samples are subject to stringent quality control protocols, including integrity analysis and analysis of nucleic acid concentration and removal of impurities.
- In silico sex determination from sequencing data.
- We perform de novo assembly for reference-free genomes.
- Available on the NovaSeq 6000 sequencer (up to 28 complete human genomes per flow cell, 90 Gb/genome, >30x, 2×150 bp). Possibility to design experiments per flow cell and per lane.
- Low coverage whole genome sequencing (lcWGS) will soon be available on both second and third generation sequencers.
In the case of non-human WGS, please note that we can customise the preparation and sequencing of the organism of interest. Please contact us without obligation. Other species: viruses, bacteria, vascular plants, other mammals, etc.
2.2. Whole human exome sequencing (WES)
- Library preparation: Illumina DNA Prep with Exon 2.5 Enrichment.
- We also have experience in using the Agilent SureSelect HS2 XT (v8) exome capture kit and in adapting protocols to other insert size options.
- Available on the NovaSeq 6000 sequencer for large projects (up to 200 exomes at 100x with 2 x 100 bp reads). Possibility to design experiments per flow cell and per lane.
- Available on the NextSeq 550 sequencer for pilot projects and projects with a small number of samples or urgent projects (6 exomes at 100x with 2 x 75 bp reads).
- Our team has processed several thousand exomes from various national and foreign research centres and health institutions.
2.3. RNA Sequencing
- mRNA
- We used oligo-dT beads to enrich the polyadenylated RNA molecules in the sample.
- Sequencing is available on the NextSeq 550 and NovaSeq 6000, with read lengths of 75 bp and 100 bp in paired-end mode, respectively.
- Library preparation protocols preserve chain information.
- Library preparation: Illumina TruSeq Stranded mRNA.
- Total RNA
- Total RNA allows the study of protein-coding RNA as well as long non-coding RNAs.
- Available read lengths are 75 bp and 100 bp in paired-end mode.
- Depletion of ribosomes in the library preparation minimises the presence of rRNA.
- Library preparation: Illumina Stranded Total RNA with Ribo-zero.
- sRNA
- Small RNA provides the ability to study RNA from 17 to 37 bp.
- Study of a wide variety of classes of regulatory RNAs.
- Library preparation: Illumina TruSeq Small RNA.
2.4. Metagenomic or shotgun sequencing
- Of interest for the creation of random fragments of sample DNA, e.g. from a clinical or environmental sample, which are subsequently sequenced and assembled to reconstruct the complete sequence.
- Library preparation: Illumina Nextera XT and various options for long readings.
- Available on the MiSeq, NextSeq 550 and NovaSeq 6000 platforms. Also available on nanopore-based sequencers (MinION Mk1B/Mk1C and GridION x5).
- Possibility to design experiments per flow cell and per line.
2.5. ChIP-Seq
- Of interest to identify DNA-associated protein binding sites in the genome. This technique combines chromatin immunoprecipitation (ChIP) with massive DNA sequencing to provide a detailed picture of protein-DNA interaction.
- Library preparation: Illumina TruSeq ChIP Library Preparation Kit.
2.6. Methyl-Seq
- Of interest to identify certain epigenetic modifications (e.g. methylation) of DNA that play a role in the regulation of gene expression and other cellular processes.
- Library preparation: Twist Targeted Methylation.
- Available on NovaSeq 6000. Possibility to design experiments per flow cell and per line.
2.7. Sequencing of pathogens and microbiome
- Sequencing of whole genomes of bacteria and viruses.
- Sequencing of 16S rRNA.
- Preparation of libraries for viruses (e.g. SARS-CoV-2, influenza and respiratory syncytial virus): Illumina CovidSeq Test.
- Available on MiSeq, NextSeq 550 and NovaSeq 6000. Ability to design experiments per flow cell and per line.
2.8. Nanopore sequencing
- Sequencing of whole genomes of bacteria and viruses.
- Sequencing of 16S rRNA.
- Adaptive sequencing (e.g. plasmid sequencing).
- Metabarcoding (e.g. IOC sequencing).
- Library preparation: consult ONT alternatives.
- Available in low performance platforms (MinION Mk1B/Mk1C) and larger scale projects (GridION x5).
3. Bioinformatics services
ITER’s Genomics Service has fully automated internal bioinformatics workflows or pipelines specific to each service, considerably reducing the time it takes to generate results.
3.1. Bioinformatics analysis of the whole genome and whole exome of humans
- Generation of sequences in FASTQ format.
- Alignment and mapping to various reference genomes (hg19, GRCh38, T2T).
- Generation of mapped sequences in BAM format.
- SNV and Indels identifation (<50 pb).
- Identification of structural variation (SV, CNV, STR, etc.) for both WGS and WES.
- Generation of data in VCF format.
- Functional annotation of the identified genetic variation.
- Possibility to use DRAGEN server applications.
3.2. Other bioinformatics analyses offered:
- Differential gene expression analysis.
- Functional annotation.
- Discovery of fusion genes.
- Mitogenome sequencing analysis.
- Microbiome sequencing analysis.
- Epigenomic analysis, etc.
3.3. Computing and storage
The HPC infrastructure (https://teidehpc.iter.es/infraestructura/) enables completely secure access, storage and data transfer. Hosted at the Neutral Access Point (NAP) and associated with the D-Alix project, it offers high availability and security 365x7x24 for power supply (TIER IV) and climate control (TIER III+) (https://www.iter.es/wp-content/uploads/2015/12/Folleto-D-ALiX.pdf). Detailed information and documentation to facilitate the connection to the HPC infrastructure can be found here (https://doc.hpc.iter.es/). Please contact our team.
4. Access to services.
4.1. Procedure.
The steps to follow to obtain information about a service or to get a quote for your project are as follows:
- The customer contacts the Genomics Service via e-mail (a ‘genomica’ [en] ‘iter.es’) to request information. The Genomics Service will send three documents: a user registration document, a document to reflect the technical characteristics of the sequencing project and the formal request for a quote. The customer will send this duly completed documentation by e-mail (Figure 3).
- The Genomics Service, in view of the documentation submitted by the client, will issue its estimate.
- If the client is in agreement with the budget, he must return the signed document and make the payment of 50% of the service in advance.
- The service will start running as soon as the samples associated with the request reach the Genomics Service.
- The Genomics Service will inform the client who must pay the remaining 50% of the service once the data and results of the service are generated and made available to the client.
- Finally, the customer will have, as a general rule, one (1) month to download the data and results of the service.
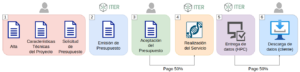
Figure 3. Flowchart describing the steps required for accessing the service and downloading data.
4.2. Access to sequence data and results of bioinformatic analyses.
Once the service is completed and the client has been informed of the availability of project data and results, access to the supercomputing infrastructure will be made available for download with appropriate security measures. The procedure is detailed in section 6.11 of this website and in the teideHPC access documentation available at https://doc.hpc.iter.es. The Genomics Service will accompany you along with the ITER teideHPC Support Team throughout the process to obtain the access credentials and configure your FTP client to download the data.
5. High-performance proteomics services.
Under development. Please contact us for availability in the coming months.

