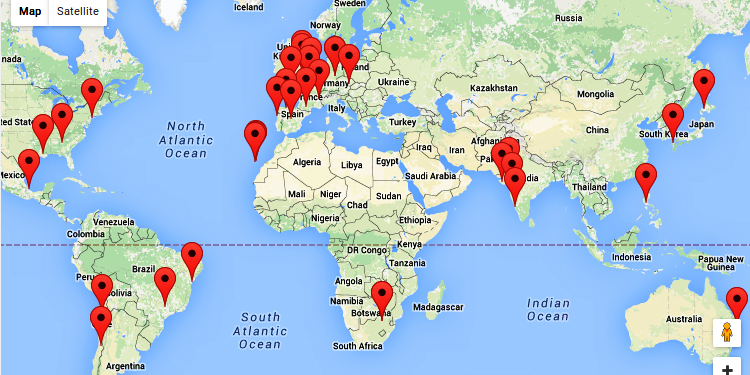
Since its public release on 28 of November of 2014, that means less than a year online, IonGAP has been used for running 300 projects.
Researchers from more than thirty institutions distributed in more than 15 countries have tested and used IonGAP platform in their studies. The results of those works have been published in scientific journals.
The roadmap of IonGAP will include several improvements. As an example, the addition of digital normalisation is one of the first new capacities that will be implemented. This new option will do all the previous quality filtering, trimming, etc. that will prepare the data for the subsequent assembly and analysis processes.
IonGAP is a publicly available integrated pipeline designed for the assembly and subsequent analysis of Ion Torrent bacterial sequence data. Both its components and their configuration are based on a research process aimed to discover the optimal combination of tools for obtaining good results from single-end reads generated by the Ion Torrent PGM sequencer.
Currently, IonGAP is a joint project between the Instituto Tecnológico y de Energías Renovables (ITER, Institute of Technology and Renewable Energies), the Universidad de La Laguna, and the Servicio Canario de la Salud (Canary Health Service). All these entities are based in Tenerife, Canary Islands, Spain. The whole platform runs on ITER’s Teide-HPC, which is the second most powerful computer in Spain at present.